this is my INCAR POSCAR KPOINTS
INCAR
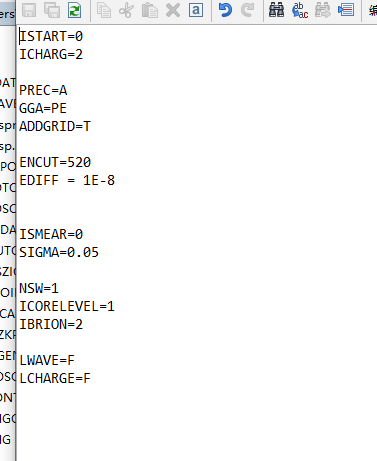
POSCAR
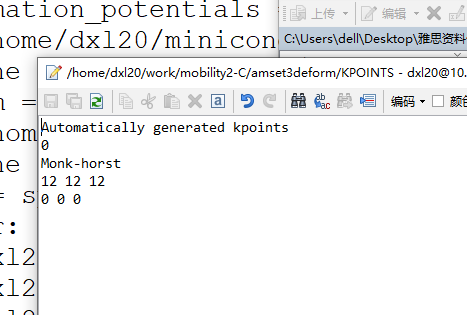
KPOINTS
Hi, I also faced this problem. In the calculation of deformation potential, I can get the deformation.h5 file using K-mesh of 9×9×3. But, when the K-mesh increased up to 13×13×4, an error has been encountered. The output of amset as follow:
Reading bulk (undeformed) calculation
Reading deformation calculation in def-01
Reading deformation calculation in def-02
Reading deformation calculation in def-03
Reading deformation calculation in def-04
Reading deformation calculation in def-05
Reading deformation calculation in def-06
unicodeify_spacegroup is deprecated
These methods have been deprecated in favor of using the Stringify mix-in class, which provides to_latex_string, to_unicode_string, etc. They will be removed in v2022.
latexify_spacegroup is deprecated
These methods have been deprecated in favor of using the Stringify mix-in class, which provides to_latex_string, to_unicode_string, etc. They will be removed in v2022.
Spacegroup: I4/mmm (139)
Found 6 strains:
- [ 0.005 0.000 0.000 0.000 0.000 0.000]
- [-0.005 0.000 0.000 0.000 0.000 0.000]
- [ 0.000 0.000 0.005 0.000 0.000 0.000]
- [ 0.000 0.000 -0.005 0.000 0.000 0.000]
- [ 0.000 0.000 0.000 0.005 0.000 0.000]
- [ 0.000 0.000 0.000 0.000 0.000 0.005]
Traceback (most recent call last):
File "/home/csnsphys/software/amset/bin/amset", line 10, in <module>
sys.exit(cli())
File "/home/csnsphys/software/amset/lib/python3.9/site-packages/click/core.py", line 829, in __call__
return self.main(*args, **kwargs)
File "/home/csnsphys/software/amset/lib/python3.9/site-packages/click/core.py", line 782, in main
rv = self.invoke(ctx)
File "/home/csnsphys/software/amset/lib/python3.9/site-packages/click/core.py", line 1259, in invoke
return _process_result(sub_ctx.command.invoke(sub_ctx))
File "/home/csnsphys/software/amset/lib/python3.9/site-packages/click/core.py", line 1259, in invoke
return _process_result(sub_ctx.command.invoke(sub_ctx))
File "/home/csnsphys/software/amset/lib/python3.9/site-packages/click/core.py", line 1066, in invoke
return ctx.invoke(self.callback, **ctx.params)
File "/home/csnsphys/software/amset/lib/python3.9/site-packages/click/core.py", line 610, in invoke
return callback(*args, **kwargs)
File "/home/csnsphys/software/amset/lib/python3.9/site-packages/amset/tools/deformation.py", line 193, in read
strain_mapping = get_symmetrized_strain_mapping(
File "/home/csnsphys/software/amset/lib/python3.9/site-packages/amset/deformation/potentials.py", line 74, in get_symmetrized_strain_mapping
calc["bandstructure"] = expand_bandstructure(
File "/home/csnsphys/software/amset/lib/python3.9/site-packages/amset/electronic_structure/symmetry.py", line 206, in expand_bandstructure
full_kpoints, _, _, _, _, kp_mapping = expand_kpoints(
File "/home/csnsphys/software/amset/lib/python3.9/site-packages/amset/electronic_structure/symmetry.py", line 157, in expand_kpoints
raise ValueError("Expected {} points but found {}".format(n_expected, n_mapped))
ValueError: Expected 676 points but found 624
Please help me to resolve this error.
I am also facing same issue for my system.I tried reducing k points.Even I tried using primitive and conventional unit cell .Everytime I am getting same problem.
@LLei It seems like your calculations have different numbers of bands. Can you check this?
@malonestar and @SUBHAJIT_SAU you need to make sure the space group matches the spacegroup found by VASP. You can do this by changing the symprec parameter. If this doesn’t work, please send me your vasprun.xml files for the bulk and displacements and I will take a look.
@alex Thanks for your reply.I will check by changing symprec parameter and get back to you.
Hi Subhajit
Are you able to overcome this error? I am still struggling with recurrent error.
No Zeeshan.
Please send me your vaspruns and OUTCARs for the bulk and deformed structures and I can take a look!