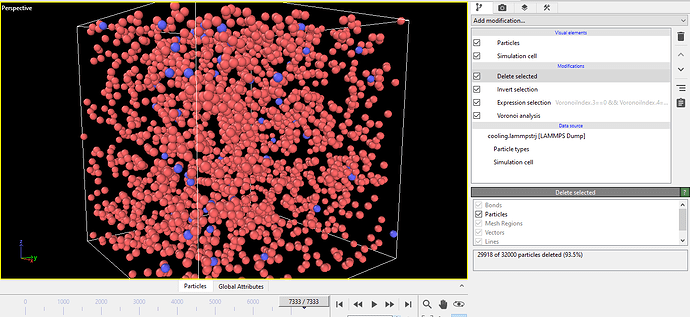
Hi OVITO Team,
I hope you are well.
Please, could you provide me with a Python script to reproduce the structures and distances shown in the attached image (1-atom to 4-atom icosahedral connections)?
I tried with Voronoi analysis, expression selection, invert, and delete tools, but I couldn’t get this result.
I’m looking for a script that can detect icosahedral clusters (like <0,0,12,0>), find their connections, and show the distances between them.
Thank you very much for your help!
Best regards,
Saad
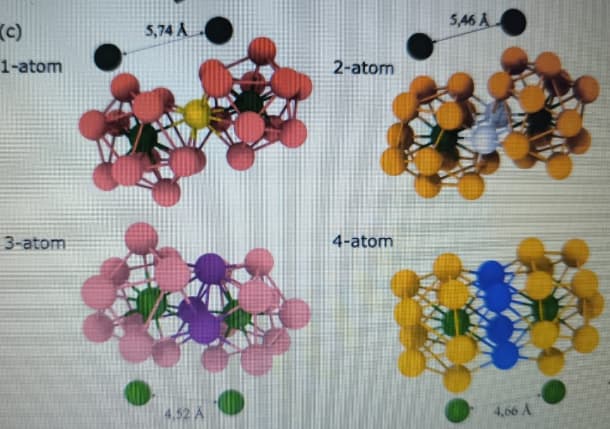
Dear team ovito; @kalcher , @stukowski
someone help in this:The cluster structures?
Thank you very much for your help!
Let’s do it step by step. At which point are you stuck?
My suggestion is that you first use the graphical version of OVITO to find out how to:
- Calculate the Voronoi indices of all atoms with the Voronoi Analysis modifier,
- Select all <0,0,12,0> atoms with the Expression Selection modifier,
- Hide all other atoms using the Invert Selection and Delete Selected modifiers.
Once you have successfully accomplished these steps, we can discuss about how to measure the distances between the <0,0,12,0> atoms.
Please note that the images in your screenshot were not created using OVITO alone. The arrows and distance annotations were probably added in an external graphics program.
1 Like
Dear Team OVITO,
First, I would like to thank Dr. @stukowski for the clear explanation and for taking the time to help. I followed the steps as described, and it gave me the result shown in the attached picture.
Could you please confirm if this is correct? And if possible, could you provide a Python script that could help automate this? I’m still a beginner, so any additional guidance would be greatly appreciated.
Thank you in advance
Best regards,
SAAD
Yes, I think you did that correctly. I would now do the following to get one step closer to the image you showed:
- Open the Voronoi Analysis modifier and activate the option “Neighbor bonds”.
- Insert an Expand Selection modifier between the Expression Selection and Invert Selection modifiers in the pipeline. In this modifier, select “… bonded to a selected particle” (3rd mode).
This should make the direct neighbors of the <0,0,12,0> atoms visible.
1 Like
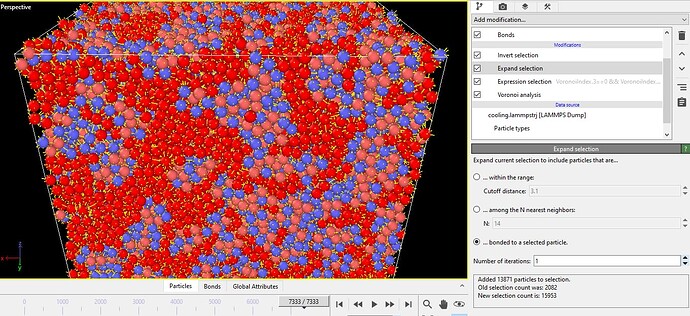
Dear Team OVITO / Dr. @stukowski ,Dr. @utt ,
OK, thank you very much for your support. I tried the method, but it gave me the result shown in the picture below.
If possible, could you please help me understand what went wrong, or guide me using the picture I attached?
Thanks in advance for your time and help.
This could be a correct outcome. Your previous screenshot showed that 6.5% of atoms in your system are <0,0,12,0> atoms. Each of them has 12 neighbor atoms. If we display these neighbors too, in addition to the central atoms, almost all atoms are shown and you get something that looks quite dense. Do you agree?
1 Like
Yes, I fully agree with you. However, I still haven’t been able to achieve the same level of connectivity as shown in the first image. So, if possible, could you please demonstrate the full process with an image and step-by-step instructions?
Thank you in advance for your time and for the helpful clarification.
Before we think about the next steps, could you please specify more precisely what your ultimate goal is?
Do you want to build a list of all pairs of connected icosahedral clusters, measure the distances between their center atoms, and classify them according to the number of atoms forming the connection? Then, what are you going to do with that comprehensive list?
1 Like
Dear Dr. @stukowski ,
Thank you for your message and for your continued support.
Yes, my goal is to calculate the Voronoi Polyhedra of specific cluster types, particularly the <0,0,12,0> (ICO-like) and <0,4,4,6> clusters, and analyze their atomic environments in Pd₁₀₀₋ₓSiₓ systems (with x = 10, 20, 30, …).
In addition, I aim to extract and analyze the linkages between these clusters — specifically the vertex-sharing (VS), edge-sharing (ES), face-sharing (FS), and interpenetrating-sharing (IS) connections — and to determine the average distances between their central atoms, as shown on the right side of the reference figure.
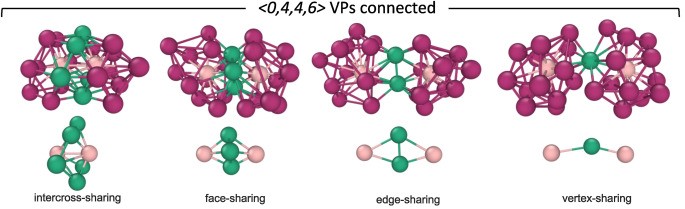
Ultimately, I want to build a list that details these pairwise connections between clusters and classify them based on the type of linkage and number of connecting atoms. This will help me better understand the medium-range order and spatial organization of these structural motifs in Pd-Si metallic glasses.
Thank you again, and I really appreciate your guidance.
Best regards,
SAAD
Dear Ovito Team, Dr @stukowski , Dr @utt , Dr @kalcher
I hope this message finds you well.
I am currently facing an issue and would greatly appreciate your assistance in resolving it. If possible, I would prefer a Python script solution. Additionally, it would be extremely helpful if the steps could be explained clearly, ideally with illustrative images.
Thank you very much in advance for your time and support.
Best regards,
SAAD