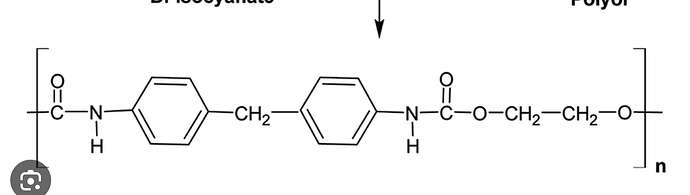
Hey I am trying to run a program to get initial configuration for polyurethane, but it is not working I don’t know what I am doing wrong or if I need to change something which I doubt, for other structures I get what I need here is my code
pressure 1.0
lammps true
ITEM END
ITEM GROUPS
polyurethane CCOC(=O)Nc2ccc(Cc1ccc(NC(=O)OCC)cc1)cc2,1,polyurethane:2
terma *C,1,polyurethane:1
termb *C,1,polyurethane:2
ITEM END
ITEM CLUSTERS
polymer block,50
ITEM END
ITEM POLYMERS
polymer
1 polyurethane,10,terma,1,termb,1
ITEM END
theses are the error I am finding
-MacBook-Pro butene16 % emc_macos build.emc
(* EMC: Enhanced Monte Carlo simulations *)
version 9.4.4, build Aug 8 2023 07:32:32, date Mon Feb 12 00:42:58 2024
valid until Jul 31, 2024
Info: script v1.0 started at Mon Feb 12 00:42:58 2024
Info: variables = {seed → -1, ntotal → 10000, fshape → 1, output →
“butene16”, field → “pcff/pcff”, location1 →
“/Applications/v9.4.4/field/”, nav → 0.6022141179, temperature →
300, radius → 5, nrelax → 100, weight_nonbond → 0.0001, weight_bond
→ 0.0001, weight_focus → 1, cutoff → 9.5, charge_cutoff → 9.5,
kappa → 4, density1 → 1, lprevious → 0, lphase → 0, f_polymer →
50, chem_polyurethane → “CCOC(=O)Nc2ccc(Cc1ccc(NC(=O)OCC)cc1)cc2”,
chem_terma → “CCOC(=O)Nc2ccc(Cc1ccc(NC(=O)OCC*)cc1)cc2”, chem_termb
→ “CCOC(=O)Nc2ccc(Cc1ccc(NC(=O)OCC)cc1)cc2"}
Info: output =
{detail → 3, wide → false, expand → false, math → true, reduced →
false, info → true, strict → true, warning → true, message → true,
debug → false, exit → true, ignore → false}
Info: field = {id → pcff, mode → cff, style → none, name →
{“/Applications/v9.4.4/field/pcff/pcff.frc”,
“/Applications/v9.4.4/field/pcff/pcff_templates.dat”}, compress →
false, error → true, debug → false, check → {atomistic → true,
charge → true}}
Info: importing ‘/Applications/v9.4.4/field/pcff/pcff.frc’
Warning: duplicate field.bond entry 40 [c:c] omitted.
Warning: duplicate field.bond entry 51 [c:h] omitted.
Warning: duplicate field.bond entry 62 [c:o_2] omitted.
Warning: duplicate field.bond entry 176 [c=:c=1] omitted.
Warning: duplicate field.bond entry 328 [c_1:o_1] omitted.
Warning: duplicate field.bond entry 364 [cp:cp] omitted.
Warning: duplicate field.bond entry 365 [cp:cp] omitted.
Warning: duplicate field.bond entry 367 [cp:h] omitted.
Warning: duplicate field.bond entry 689 [o:p] omitted.
Info: groups = {ngroups → 3, group → {
{id → polyurethane, chemistry →
“CCOC(=O)Nc2ccc(Cc1ccc(NC(=O)OCC)cc1)cc2”, depth → 8, charges
→ forcefield, charge → 0, terminator → false, nconnects → 4,
connects → {
{source → $end1, destination → {index → polyurethane, site →
$end2}},
{source → $end1, destination → {index → terma, site →
$end1}},
{source → $end2, destination → {index → polyurethane, site →
$end1}},
{source → $end2, destination → {index → termb, site →
$end1}}}},
{id → terma, chemistry →
"CCOC(=O)Nc2ccc(Cc1ccc(NC(=O)OCC)cc1)cc2”, depth → 8, charges →
forcefield, charge → 0, terminator → false, nconnects → 1,
connects →
{source → $end1, destination → {index → polyurethane, site →
$end1}}},
{id → termb, chemistry →
“*CCOC(=O)Nc2ccc(Cc1ccc(NC(=O)OCC)cc1)cc2”, depth → 8, charges →
forcefield, charge → 0, terminator → false, nconnects → 1,
connects →
{source → $end1, destination → {index → polyurethane, site →
$end2}}}}, ndeletes → 0, npolymers → 0}
Info: field = {mode → apply, style → none, error → true, debug → false,
check → {atomistic → true, charge → true}, increment → warn}
Info: applying ‘/Applications/v9.4.4/field/pcff/pcff.frc’
Warning: increment pair {o_2, c-} set to zero.
Warning: increment pair {c-, na} set to zero.
Info: variables = {lg_polyurethane → 45, lg_terma → 46, lg_termb → 46,
norm_polymer → 1, l_polymer → 542, mg_polyurethane → 340.38225,
mg_terma → 341.39022, mg_termb → 341.39022, m_polymer → 4086.60294,
n_polymer → 50, ntotal → 0, mtotal → 0}
Info: simulation = {units → {permittivity → 1, seed → seed}, types → {
coulomb → {pair → {active → true, cutoff → charge_cutoff}}}}
Info: clusters = {progress → none, nbodys → 0, nclusters → 0, ngrafts → 0,
npolymers → 1,
polymer →
{id → polymer, system → main, type → block, niterations → 1000, n
→ 50, groups → {polyurethane, terma, termb}, weights → {1, 1, 1},
nrepeat → {10, 1, 1}}}
delta = {systems → 1, bodies → 0, clusters → 50, sites → 27100}
Info: field = {mode → apply, style → none, error → true, debug → false,
check → {atomistic → true, charge → true}, increment → warn}
Info: applying ‘/Applications/v9.4.4/field/pcff/pcff.frc’
Warning: increment pair {o_2, c-} set to zero.
Warning: increment pair {c-, na} set to zero.
Warning: no torsion coefficients found for {c, c, o_2, c-}.
Warning: no torsion coefficients found for {hc, c, o_2, c-}.
Error: core/script/field/entry.c:641 ScriptFieldEntryApply:
Missing force field parameters.
Program aborted.