Dear all,
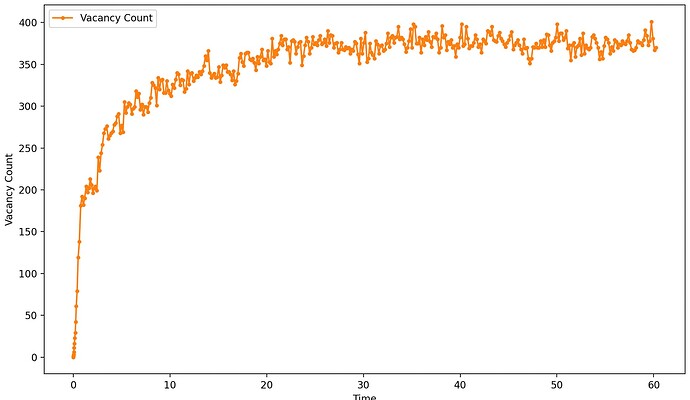
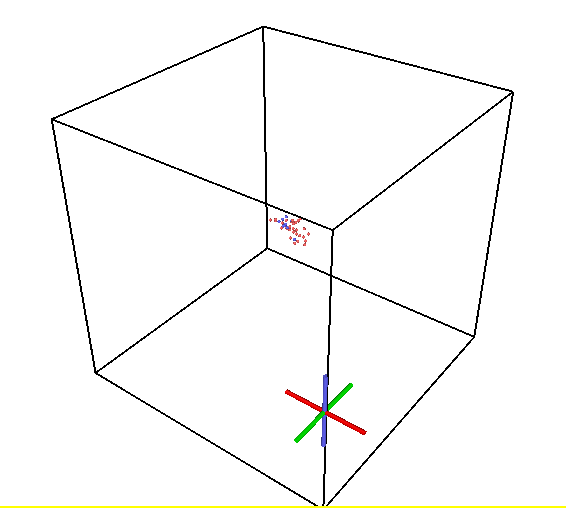
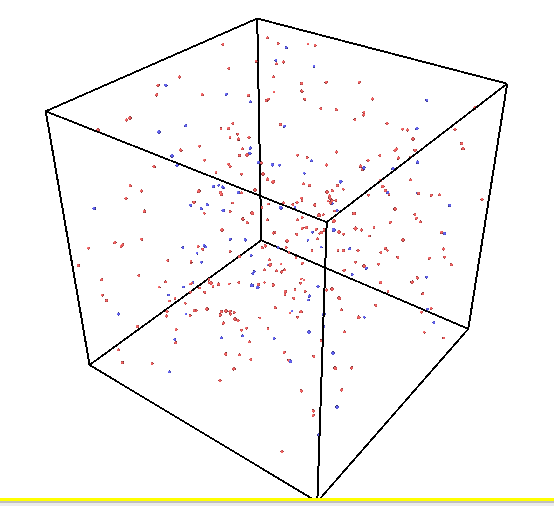
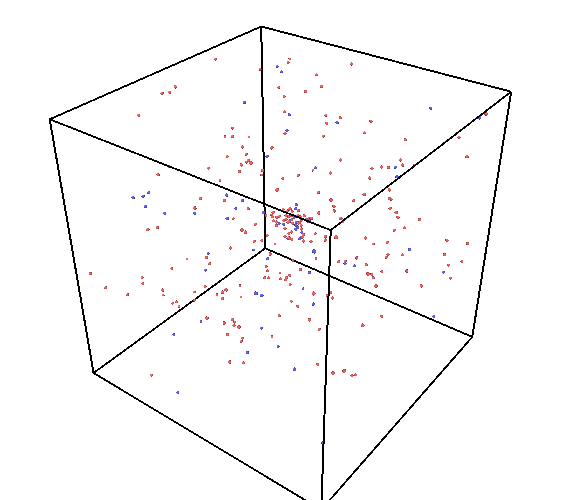
I’m investigating the cascade behaviors in U-Zr alloys at high temperatures and found unexpectedly increasing Frenkel pair numbers as attached. This behavior seems different from that reported in references for similar systems. These unexpected defects are distributed in the whole box.
I’ve tested the pure U metal at the same temperature and alloy with different phase structures at low temperatures, but such a trend is not observed in those cases.
Has anyone experienced a similar problem or have suggestions on how to address this? Thanks in advance!
Best regards.
Zhou
Details of the analysis:
Vacancy Number vs. Time: The vacancy number is tracked using OVITO with the Wigner-Seitz defect analysis, where occupancy == 0 is used to identify vacancies.
The snapshot of the vacancy distributions at 0.25 ps, 5.9 ps, and 40 ps.
The input files are attached, where relax.data is the 646464 supercell with 524288 atoms relaxed in an NPT ensemble for 20 ps. The applied potential is developed by Moore, which is MEAM for U-Zr alloy at high temperatures.
units metal
atom_style atomic
dimension 3
boundary p p p
timestep 0.0001
read_data relax.data
pair_style hybrid/overlay zbl 1.5 2.5 meam
pair_coeff 1 1 zbl 92 92
pair_coeff 1 2 zbl 92 40
pair_coeff 2 2 zbl 40 40
pair_coeff * * meam library.meam U Zr U-Zr.meam U Zr
neighbor 2.0 bin
neigh_modify delay 0 every 1 check yes
thermo 500
fix nve_fix all nve
velocity all create 1000 123456 dist gaussian
region out_region sphere 112.86 112.86 112.86 100 units box side out
group damp_atom region out_region
fix damp_fix damp_atom langevin 1000 1000 0.1 123456 zero yes
fix t_re all dt/reset 10 1.0e-6 1.0e-3 0.005 units box
group pka id 274370
velocity pka set 48.1 144.4 240.6 units lattice
compute pka_ke pka ke/atom
compute pka_totalke pka reduce sum c_pka_ke
thermo_style custom step time temp etotal pe ke c_pka_totalke
dump 3 all custom 500 pka.xyz id type x y z
dump_modify 3 time yes
dump 4 pka custom 1 pka.dump id type x y z
dump_modify 4 time yes
run 200000
Log file attached.
LAMMPS (29 Aug 2024 - Update 1)
Reading data file …
orthogonal box = (-2.8737387 -2.8737387 -2.8737387) to (222.78542 222.78542 222.78542)
2 by 4 by 5 MPI processor grid
reading atoms …
524288 atoms
reading velocities …
524288 velocities
read_data CPU = 7.149 seconds
333205 atoms in group damp_atom
1 atoms in group pka
CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE
Your simulation uses code contributions which should be cited:
- Type Label Framework: https://doi.org/10.1021/acs.jpcb.3c08419
The log file lists these citations in BibTeX format.
CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE
Neighbor list info …
update: every = 1 steps, delay = 0 steps, check = yes
max neighbors/atom: 2000, page size: 100000
master list distance cutoff = 7.5
ghost atom cutoff = 7.5
binsize = 3.75, bins = 61 61 61
2 neighbor lists, perpetual/occasional/extra = 2 0 0
(1) pair meam, perpetual
attributes: full, newton on
pair build: full/bin/atomonly
stencil: full/bin/3d
bin: standard
(2) pair meam, perpetual, half/full from (1)
attributes: half, newton on
pair build: halffull/newton
stencil: none
bin: none
Setting up Verlet run …
Unit style : metal
Current step : 0
Time step : 0.0001
Per MPI rank memory allocation (min/avg/max) = 34.45 | 34.68 | 34.83 Mbytes
Step Time Temp TotEng PotEng KinEng c_pka_totalke
0 0 1014.7474 -2692948.9 -2761717.7 68768.835 999.79564
500 0.0093391492 1023.9814 -2692994.2 -2762388.8 69394.618 719.65762
1000 0.01972648 1053.0807 -2693135.8 -2764502.5 71366.665 685.94248
1500 0.03126568 1085.4644 -2693647.2 -2767208.5 73561.285 506.624
2000 0.046300864 1108.8236 -2694628.8 -2769773.2 75144.322 236.62272
2500 0.062409104 1111.1433 -2695766.9 -2771068.4 75301.53 165.35197
3000 0.088495138 1098.9632 -2697314.7 -2771790.8 74476.092 66.866681
3500 0.11990297 1091.8592 -2698679.5 -2772674.1 73994.656 13.161264
4000 0.15747082 1082.0045 -2700061.4 -2773388.2 73326.81 5.3569883
4500 0.20512725 1076.2034 -2701276.5 -2774210.2 72933.676 4.7677699
5000 0.25507073 1071.0974 -2702153.8 -2774741.4 72587.639 1.4595814
5500 0.31063707 1062.3593 -2702806.3 -2774801.7 71995.466 0.57230149
6000 0.40282232 1059.1798 -2703518.3 -2775298.3 71779.993 0.6198328
6500 0.50509537 1054.8888 -2704080.7 -2775569.9 71489.198 0.5896941
…
194000 58.338537 1009.5757 -2712258.3 -2780676.6 68418.354 0.06411991
194500 58.499201 1012.771 -2712117.9 -2780752.8 68634.9 0.084909225
195000 58.658136 1012.4902 -2712080.7 -2780696.6 68615.871 0.10513696
195500 58.814055 1012.4752 -2712218.8 -2780833.7 68614.853 0.13331323
196000 58.974459 1011.0663 -2712153.1 -2780672.5 68519.37 0.1112543
196500 59.130525 1012.9537 -2711982.3 -2780629.5 68647.28 0.14530792
197000 59.290154 1012.5529 -2712039.6 -2780659.7 68620.119 0.055896908
197500 59.448266 1012.8424 -2712043 -2780682.8 68639.735 0.21904679
198000 59.605765 1013.1939 -2712098 -2780761.5 68663.558 0.049729567
198500 59.766217 1011.1245 -2712207.9 -2780731.2 68523.317 0.086338604
199000 59.924759 1011.9659 -2712276.1 -2780856.4 68580.333 0.079297101
199500 60.080352 1011.4277 -2712267.7 -2780811.6 68543.865 0.20088287
200000 60.237456 1011.9751 -2712078.4 -2780659.3 68580.962 0.070622574
Loop time of 72511.5 on 40 procs for 200000 steps with 524288 atoms
Performance: 0.072 ns/day, 333.006 hours/ns, 2.758 timesteps/s, 1.446 Matom-step/s
98.4% CPU use with 40 MPI tasks x no OpenMP threads
MPI task timing breakdown:
Section | min time | avg time | max time |%varavg| %total
Pair | 70106 | 70765 | 71301 | 123.2 | 97.59
Neigh | 52.843 | 56.825 | 61.07 | 31.8 | 0.08
Comm | 354.76 | 869.87 | 1557.9 |1122.2 | 1.20
Output | 45.901 | 75.595 | 119.15 | 209.3 | 0.10
Modify | 414.67 | 482.88 | 521.9 | 104.7 | 0.67
Other | | 261.2 | | | 0.36
Nlocal: 13107.2 ave 13353 max 12772 min
Histogram: 1 3 3 1 10 6 1 2 7 6
Nghost: 12350.5 ave 12784 max 12053 min
Histogram: 10 5 1 0 7 7 2 2 4 2
Neighs: 472201 ave 480965 max 459683 min
Histogram: 1 3 3 2 7 6 3 3 6 6
FullNghs: 944402 ave 961560 max 919200 min
Histogram: 1 3 3 1 7 7 2 3 6 7
Total # of neighbors = 37776096
Ave neighs/atom = 72.052185
Neighbor list builds = 775
Dangerous builds = 0
Total wall time: 20:08:39