Dear, Lammps uers
I am trying to calculate self diffusion coefficient from MSD.
I made 100 water molecular(liquid) 1 atm, 25 C.
First of all, I gave time 100ps to equilibriate the system with npt.
I got density about 0.9 g/ml3, and 3332 angstrom^2 volume.
And next, I simulate the system using nvt. I fixed the system volume with fix deform for 10000 run.
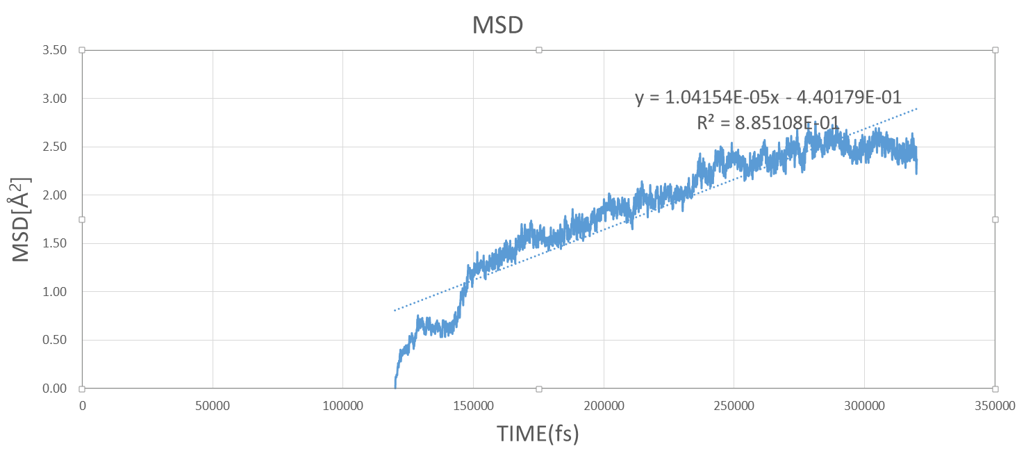
Then, I used msd command with O atoms.
But, diffusion coefficient is too low, it is 1/100 than I expected.
I ran the simulation 200ps. ( I tried much more time, but result was same)
Can any one give advises ?
Units
units real
Resion
boundary p p p
Force type
atom_style full
pair_style lj/cut/coul/cut 9 9
bond_style harmonic
angle_style harmonic
Atom definition
read_data data_100W.txt
velocity all create 300 8343 dist gaussian units box
Neighbor list
neighbor 2.0 bin
neigh_modify every 10 delay 0 check yes
Calculation
thermo_style custom step pe ke etotal temp press vol density
thermo 10
Minimization
minimize 1.0e-5 1.0e-7 1000 100000
Timestep
timestep 1
Fix molecular
fix 1 all shake 1.0e-4 10 0 b 1 a 1
Dynamic type
fix 2 all npt temp 298.15 298.15 100 iso 1 1 100
Variable
variable fs equal 1*step
variable density equal “density”
variable vol equal “vol”
Print density
fix 3 all print 10 “{fs} {density}” file fsDensity.txt screen no
fix 4 all print 10 “{fs} {vol}” file fsVol.txt screen no
Start running
run 100000
Save the restart file
write_restart restart100W.txt
Units
units real
Resion
boundary p p p
Force type
atom_style full
pair_style lj/cut/coul/cut 9 12
bond_style harmonic
angle_style harmonic
Atom definition
read_restart restart100W.txt
Neighbor list
neighbor 2.0 bin
neigh_modify every 10 delay 0 check yes
Calculation
thermo_style custom step pe ke etotal temp press vol density lx ly lz
thermo 100
Timestep
timestep 1
Fix molecular
fix 1 all shake 1.0e-4 10 0 b 1 a 1
Dynamic type
fix 2 all deform 1 x final 0.0 14.937 y final 0.0 14.937 z final 0.0 14.937
fix 3 all nvt temp 298.15 298.15 100
run 10000
unfix 2
run 10000
Group
group ox type 1
Compute
compute 1 ox msd
compute 5 ox vacf
Variable
variable msd equal c_1[4]
variable vacf equal c_5[4]
variable fs equal 1*step
variable Temp equal “temp”
variable Vol equal “vol”
variable density equal “density”
Print results and make files
fix 7 all print 10 “{fs} {msd}” file msd.txt screen no
fix 9 all print 10 “{fs} {vacf}” file vacf.txt screen no
fix 8 all vector 1 c_5[4]
variable diff equal dt*trap(f_8)
fix 10 all print 10 “{fs} {diff}” file diff.txt screen no
Dump
dump 1 all xyz 100 dump.xyz
dump_modify 1 element O H C
Run a simulation
run 200000