jwjeffr
September 12, 2023, 10:14pm
1
Hello,
When following the example for the dynamical_matrix command (here ) and turning on the minimization flag, there are substantial negative values. Here are the first six lines of the output file that’s generated when I run it:
5407.73126648 0.60396688 0.00000175
This shouldn’t be the case for a minimized configuration, right? Shouldn’t the elements of the Hessian should be positive at a minimum? The matrix’s eigenvalues are negative as well (using this tool ). The first 8 are:
8562.748779308313
I have tried the command on other systems as well, and there have been negative values in every case. I’m not really sure what to make of this.
Perhaps @charlessievers can comment on this.
@jwjeffr try running these python commands on the dynamical matrix produced by the example.
python
>>> import numpy as np
>>> dynmat = np.loadtxt("dynmat.dat")
>>> dynlen = int(3*np.sqrt(len(dynmat)/3))
>>> dynmat = dynmat.reshape((dynlen, dynlen))
>>> eigvals, eigvecs = np.linalg.eig(dynmat)
>>> print(eigvals)
I would expect the following eigenvalues.
[ 1.02087855e+04 1.02086846e+04 1.02086687e+04 -7.02565700e-07
1.09978518e-08 -2.14679921e-07 8.76037508e+03 8.76609157e+03
8.77985671e+03 8.77421085e+03 8.76454125e+03 8.77580396e+03
5.87442129e+03 5.87183508e+03 5.87701407e+03 5.87961102e+03
5.87961998e+03 5.87961593e+03 1.87765175e+03 1.87697734e+03
1.87749573e+03 1.87713498e+03 1.87747312e+03 1.87715744e+03]
Hey Charles,
Thanks for the reply. I’m getting very different values, both with and without minimization.
With minimization off, I’m getting:
[18291.47498894 +0.j 16826.16995723 +0.j
15052.74248748 +0.j 13481.35824052 +0.j
11993.33955647 +0.j 11738.41797648 +0.j
10437.00465851 +0.j -99.24306209 +81.19351375j
-99.24306209 -81.19351375j 239.01553747 +0.j
9301.07339057 +0.j 8703.29381758 +0.j
7672.39998404 +0.j 6490.2455242 +459.03829328j
6490.2455242 -459.03829328j 6425.47294151 +0.j
5512.28512477 +0.j 3875.19846074 +0.j
1830.80372983 +0.j 2556.33288212 +0.j
2155.9857413 +0.j 2262.19726512 +54.13875737j
2262.19726512 -54.13875737j 2268.21773236 +0.j ]
With minimization (and the dump lines commented out because I don’t have EXTRA-DUMP enabled), I’m getting:
[15773.89957411 +0.j 14891.91401907 +0.j
13447.21476291+159.09077261j 13447.21476291-159.09077261j
12330.48643728 +0.j -1157.881718 +0.j
11130.77015491 +0.j 10679.91571791 +0.j
10360.31082596 +0.j 8573.77137431+649.09068966j
8573.77137431-649.09068966j 7752.65122494 +0.j
6661.62083014 +0.j 6266.26994726 +0.j
-97.62537959 +0.j 220.58766299 +0.j
4499.06141346 +0.j 4198.20182449 +0.j
1423.33929023 +0.j 2582.89660601 +0.j
2392.01937294 +0.j 1997.62990666 +0.j
2197.92664334 +88.48427875j 2197.92664334 -88.48427875j]
I cloned LAMMPS (origin/develop 1ebb60a33f8c13c35d40067cf02e6210b96d86f)
cd examples/PACKAGES/phonon/dynamical_matrix_command/Silicon
../../../../../build/lmp -in in.silicon
python
>>> import numpy as np
>>> dynmat = np.loadtxt("dynmat.dat")
>>> dynlen = int(3*np.sqrt(len(dynmat)/3))
>>> dynmat = dynmat.reshape((dynlen, dynlen))
>>> eigvals, eigvecs = np.linalg.eig(dynmat)
>>> print(eigvals)
and this yielded
[-6.42091335e-08+7.90277636e-08j -6.42091335e-08-7.90277636e-08j
-1.09533249e-06+0.00000000e+00j 1.02087855e+04+0.00000000e+00j
1.02086846e+04+0.00000000e+00j 1.02086687e+04+0.00000000e+00j
8.76037508e+03+0.00000000e+00j 8.76609157e+03+0.00000000e+00j
8.77985671e+03+0.00000000e+00j 8.77421085e+03+0.00000000e+00j
8.76454125e+03+0.00000000e+00j 8.77580396e+03+0.00000000e+00j
5.87442129e+03+0.00000000e+00j 5.87183508e+03+0.00000000e+00j
5.87701407e+03+0.00000000e+00j 5.87961102e+03+0.00000000e+00j
5.87961998e+03+0.00000000e+00j 5.87961593e+03+0.00000000e+00j
1.87765175e+03+0.00000000e+00j 1.87697734e+03+0.00000000e+00j
1.87749573e+03+0.00000000e+00j 1.87713498e+03+0.00000000e+00j
1.87747312e+03+0.00000000e+00j 1.87715744e+03+0.00000000e+00j]
With energy minimization (I commented out the dump commands)
[ 1.02086578e+04+0.00000000e+00j 1.02086513e+04+0.00000000e+00j
1.02086527e+04+0.00000000e+00j -1.16725621e-07+0.00000000e+00j
-1.46634440e-07+3.71726258e-07j -1.46634440e-07-3.71726258e-07j
8.76824945e+03+0.00000000e+00j 8.77207023e+03+0.00000000e+00j
8.76937614e+03+0.00000000e+00j 8.77093843e+03+0.00000000e+00j
8.76903712e+03+0.00000000e+00j 8.77128380e+03+0.00000000e+00j
5.87651234e+03+0.00000000e+00j 5.87601134e+03+0.00000000e+00j
5.87702835e+03+0.00000000e+00j 5.87754508e+03+0.00000000e+00j
5.87754627e+03+0.00000000e+00j 5.87752871e+03+0.00000000e+00j
1.87725025e+03+0.00000000e+00j 1.87738299e+03+0.00000000e+00j
1.87728193e+03+0.00000000e+00j 1.87735103e+03+0.00000000e+00j
1.87728489e+03+0.00000000e+00j 1.87734812e+03+0.00000000e+00j]
If you did the exact same, please let me know what machine/OS and lammps commit you are using.
I just re-cloned and re-built as well and it seems to be working fine now, both w/ and w/o minimization. Really weird. I guess there’s something wrong with my original build.
Thanks Charles!
@charlessievers
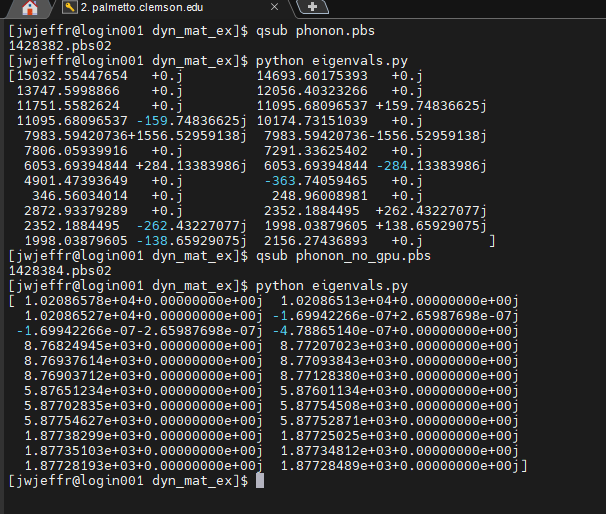
I just re-built the executable with the GPU package and it’s returning eigenvalues with large imaginary components again. Using the same executable, but without the -sf gpu flag, I am getting more reasonable values. I attached a screenshot below - these are with minimization on.
@jwjeffr is your lammps compiled with gpu single precision? If so, recompile with double precision and try again.
I was compiling with mixed precision. Switching to double worked, thank you