Hello
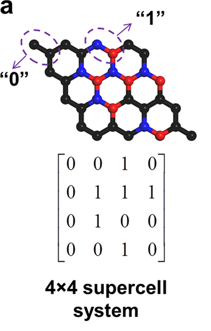
I am trying to recreate the schematic attached. I have a relaxed pwscf.in structure and I read it using pw = PWInput.from_file("pwscf.in").
I can print out the structure with pw2.structure:
Structure Summary
Lattice
abc : 9.84 9.840000000000973 16.0
angles : 90.0 90.0 119.99999999999673
volume : 1341.6548693869058
A : 9.84 0.0 0.0
B : -4.92 8.52168997324 0.0
C : 0.0 0.0 16.0
PeriodicSite: C (1.2300, 0.7101, 8.0000) [0.1667, 0.0833, 0.5000]
PeriodicSite: C (-0.0000, 1.4203, 8.0000) [0.0833, 0.1667, 0.5000]
PeriodicSite: C (0.0000, 2.8406, 8.0000) [0.1667, 0.3333, 0.5000]
PeriodicSite: C (-1.2300, 3.5507, 8.0000) [0.0833, 0.4167, 0.5000]
PeriodicSite: C (-1.2300, 4.9710, 8.0000) [0.1667, 0.5833, 0.5000]
PeriodicSite: C (-2.4600, 5.6811, 8.0000) [0.0833, 0.6667, 0.5000]
PeriodicSite: C (-2.4600, 7.1014, 8.0000) [0.1667, 0.8333, 0.5000]
PeriodicSite: C (-3.6900, 7.8116, 8.0000) [0.0833, 0.9167, 0.5000]
PeriodicSite: C (3.6900, 0.7101, 8.0000) [0.4167, 0.0833, 0.5000]
PeriodicSite: C (2.4600, 1.4203, 8.0000) [0.3333, 0.1667, 0.5000]
PeriodicSite: C (2.4600, 2.8406, 8.0000) [0.4167, 0.3333, 0.5000]
PeriodicSite: C (1.2300, 3.5507, 8.0000) [0.3333, 0.4167, 0.5000]
PeriodicSite: C (1.2300, 4.9710, 8.0000) [0.4167, 0.5833, 0.5000]
PeriodicSite: C (-0.0000, 5.6811, 8.0000) [0.3333, 0.6667, 0.5000]
PeriodicSite: C (0.0000, 7.1014, 8.0000) [0.4167, 0.8333, 0.5000]
PeriodicSite: C (-1.2300, 7.8116, 8.0000) [0.3333, 0.9167, 0.5000]
PeriodicSite: C (6.1500, 0.7101, 8.0000) [0.6667, 0.0833, 0.5000]
PeriodicSite: C (4.9200, 1.4203, 8.0000) [0.5833, 0.1667, 0.5000]
PeriodicSite: C (4.9200, 2.8406, 8.0000) [0.6667, 0.3333, 0.5000]
PeriodicSite: C (3.6900, 3.5507, 8.0000) [0.5833, 0.4167, 0.5000]
PeriodicSite: C (3.6900, 4.9710, 8.0000) [0.6667, 0.5833, 0.5000]
PeriodicSite: C (2.4600, 5.6811, 8.0000) [0.5833, 0.6667, 0.5000]
PeriodicSite: C (2.4600, 7.1014, 8.0000) [0.6667, 0.8333, 0.5000]
PeriodicSite: C (1.2300, 7.8116, 8.0000) [0.5833, 0.9167, 0.5000]
PeriodicSite: C (8.6100, 0.7101, 8.0000) [0.9167, 0.0833, 0.5000]
PeriodicSite: C (7.3800, 1.4203, 8.0000) [0.8333, 0.1667, 0.5000]
PeriodicSite: C (7.3800, 2.8406, 8.0000) [0.9167, 0.3333, 0.5000]
PeriodicSite: C (6.1500, 3.5507, 8.0000) [0.8333, 0.4167, 0.5000]
PeriodicSite: C (6.1500, 4.9710, 8.0000) [0.9167, 0.5833, 0.5000]
PeriodicSite: C (4.9200, 5.6811, 8.0000) [0.8333, 0.6667, 0.5000]
PeriodicSite: C (4.9200, 7.1014, 8.0000) [0.9167, 0.8333, 0.5000]
PeriodicSite: C (3.6900, 7.8116, 8.0000) [0.8333, 0.9167, 0.5000]
and I am able to manipulate individually e.g. specific atom 0 with pw.structure[0]="B"
For now, I just generate randomly a 1D-array of 0s and 1s and change those values according to
generated = np.random.choice([0, 1], size=(32,), p=[0.8,.2])
idx, = np.where(generated == 1)
pw.structure[idx]="B"
However, when I run pw.write_file('pw01.in') to write the transformed input file, all the atoms are changed to B with pseudopotential mismatch:
ATOMIC_SPECIES
B1 10.8110 C.pbesol-n-kjpaw_psl.1.0.0.UPF
ATOMIC_POSITIONS crystal
B1 0.166667 0.083333 0.500000
B1 0.083333 0.166667 0.500000
B1 0.166667 0.333333 0.500000
B1 0.083333 0.416667 0.500000
B1 0.166667 0.583333 0.500000
B1 0.083333 0.666667 0.500000
B1 0.166667 0.833333 0.500000
B1 0.083333 0.916667 0.500000
B1 0.416667 0.083333 0.500000
B1 0.333333 0.166667 0.500000
B1 0.416667 0.333333 0.500000
B1 0.333333 0.416667 0.500000
B1 0.416667 0.583333 0.500000
B1 0.333333 0.666667 0.500000
B1 0.416667 0.833333 0.500000
B1 0.333333 0.916667 0.500000
B1 0.666667 0.083333 0.500000
B1 0.583333 0.166667 0.500000
B1 0.666667 0.333333 0.500000
B1 0.583333 0.416667 0.500000
B1 0.666667 0.583333 0.500000
B1 0.583333 0.666667 0.500000
B1 0.666667 0.833333 0.500000
B1 0.583333 0.916667 0.500000
B1 0.916667 0.083333 0.500000
B1 0.833333 0.166667 0.500000
B1 0.916667 0.333333 0.500000
B1 0.833333 0.416667 0.500000
B1 0.916667 0.583333 0.500000
B1 0.833333 0.666667 0.500000
B1 0.916667 0.833333 0.500000
B1 0.833333 0.916667 0.500000
Additionally, pw.as_dict() shows the species and pseudopotential mismatch:
{'species': [{'element': 'B', 'occu': 1}],
'abc': [0.0833333, 0.666667, 0.5],
'xyz': [-2.460001968, 5.681129489389992, 8.0],
'label': 'B',
'properties': {'pseudo': 'C.pbesol-n-kjpaw_psl.1.0.0.UPF'}}
Does anybody know how to solve this problem?
I hope this makes sense.
Cheers,
Hud