Hi Alex,
I’ve raised a question regarding the differences in the deformation potential values with and without soc. I tried to modify the code to write out the deformation potential values for each strain component for every band. I have observed that, for spin polarized calculations, values in the normalization matrix are double as the same strain repeats for spin up and spin down bands. This issue can be resolved by separately determining normalization matrix for different spin channels.
I’m attaching all the results below. Please have a look at it and correct me if I’m wrong.
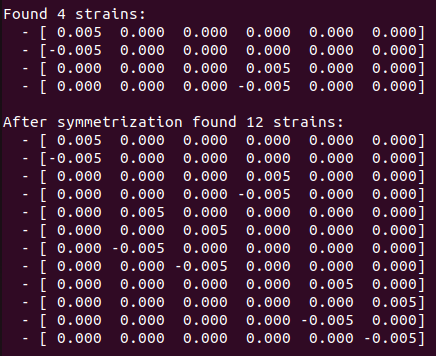
Strains for without soc calculation:
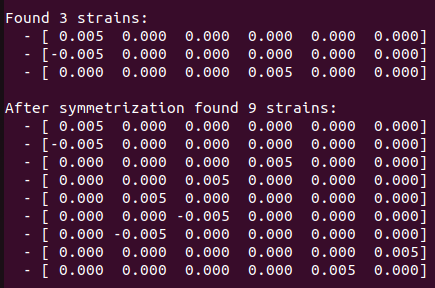
Strains for SOC calculation
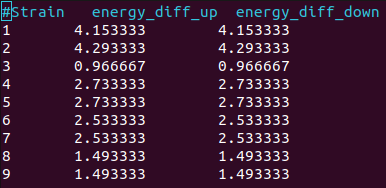
Energy differences at VBM for spin-up and spin-down bands for each strain (without soc)
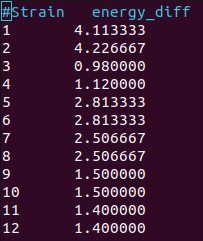
Energy differences at VBM for soc calculation (as degeneracy breaks, only one spin channel exists)
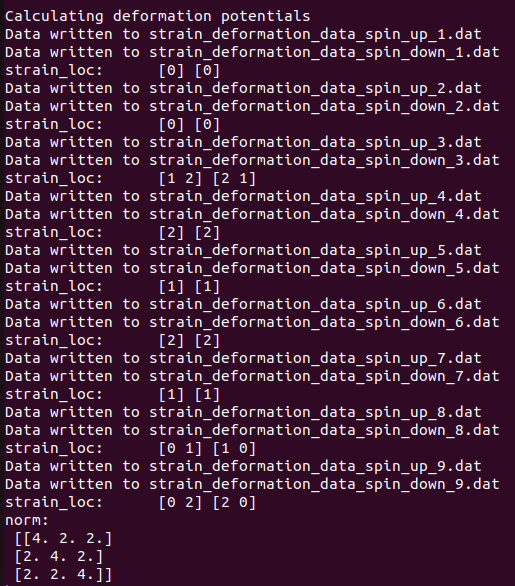
As the energy differences for positive and negative strains should be normalized, strain_loc parameter is used to record occurrences to eventually build a normalization matrix. I’ve extracted both strain_loc and norm.
strain_loc and norm for without soc case:
Here strain_loc repeats for each spin channel and therefore counted twice its actual occurrence.
Actual norm should be
[[2 1 1]
[1 2 1]
[1 1 2]]
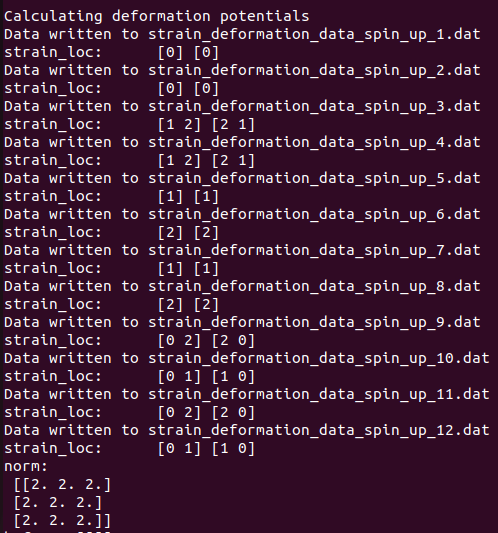
Now in soc case its counted correctly:
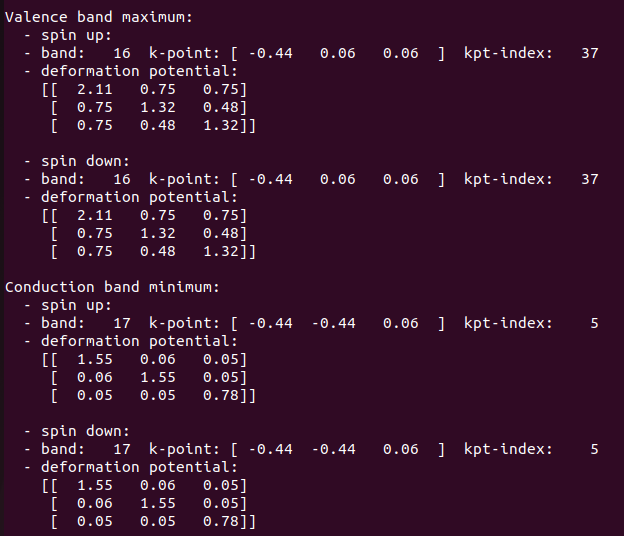
The deformation potential obtained for without soc case:
using corrected normalization matrix, the deformation potential values obtained would be double and that would be comparable to soc case.
Deformation potential values with soc case:
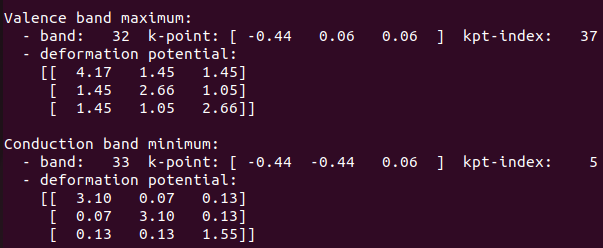
I guess this would be the case not just with and without soc case, but also, spin-polarized and non-spin polarized calculations as well.
So, I request you to go through this once and resolve this issue.
Thank you.