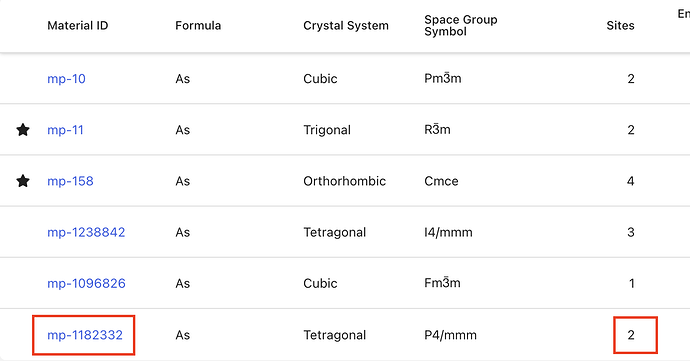
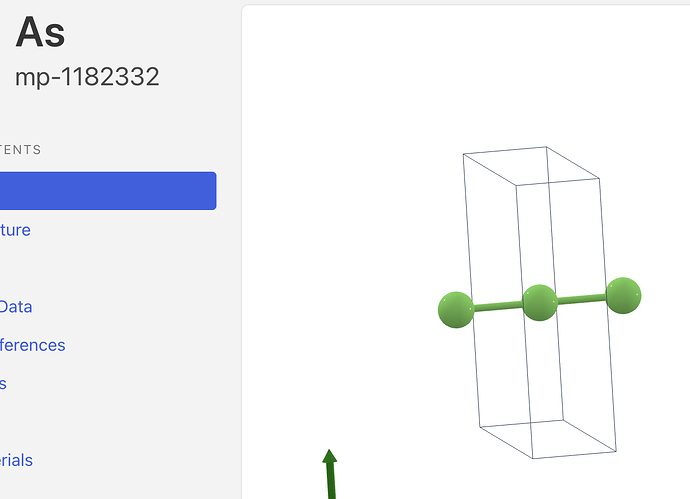
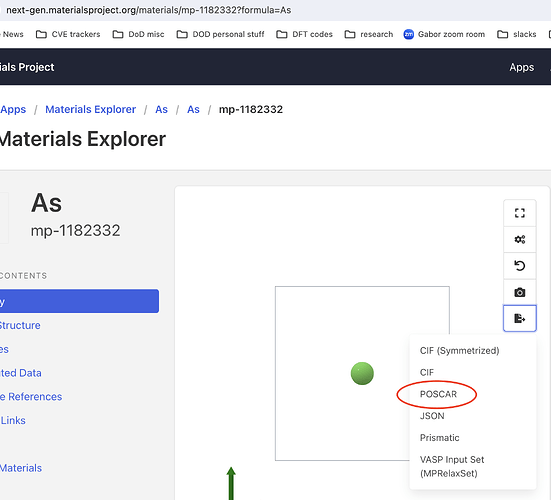
Can anyone explain why this structure https://next-gen.materialsproject.org/materials/mp-1182332?formula=As is labeled as having 2 sites (screenshots below)? Just a bug? Related to Web site structure search results table shows wrong # of sites - #4 by tschaume @tschaume ?
Hey @noam.bernstein, the structure has two sites as you can see by pulling the document / structure from the API:
from mp_api.client import MPRester
with MPRester(<your API key>) as mpr:
doc = mpr.materials.summary.search(fields=["structure"], material_ids=["mp-1182332"])[0]
print(len(doc.structure))
Which prints 2. The crystal visualization shows a periodic image.
The web site’s POSCAR export gives a 1 atom cell
As1
1.0
7.4850936655054756 0.0000000000000000 0.0000000000000005
0.0000000000000012 7.4850936655054756 0.0000000000000005
0.0000000000000000 0.0000000000000000 2.3082939867779517
As
1
direct
0.5000000000000000 0.5000000000000000 0.5000000000000000 As
The next-gen site doesn’t allow you to directly download POSCARs, you’d need to pull from the API and then write a POSCAR file from the structure (using the code snippet I sent before)
Are you using the legacy site to directly download the POSCAR? If so, keep in mind that the legacy site is frozen at an older data release (please correct me if I’m wrong @tschaume @munrojm). More, that specific material appears to be deprecated on the legacy site
I don’t think it’s the “legacy site”. I’m using https://next-gen.materialsproject.org/materials . That says “next-gen”, so it’d be weird for it to be legacy.
OK the export gives you the primitive cell, at least for POSCAR format, whereas directly downloading from the API gives you the computational cell that was used in the calculation.
@ruoxiyang and @tschaume : can we make it clearer that the export is for the primitive cell, maybe give an option for the conventional cell?
Out of curiosity, why is the calculation done on a cell that’s larger than it needs to be?
Also, the “sites” label on the main page table should possibly be more explicit that it’s conventional cell sites.
I can’t say why precisely without knowing the origin of this particular structure. Usually, structures with an experimental origin come from the ICSD or another database, and these may be any number of repeat units of the primitive cell. If the symmetry determination fails on the experimental supercell (for any of a variety of reasons), then the reduction would be to an incorrect primitive cell and not reflect the anticipated symmetry.
Another reason (not the case here) would be testing different magnetic orderings of the same structure.
hi @noam.bernstein thanks for the feedback. The number of atoms you see on the detail page is the number of atoms in a conventional cell (determined with the Pymatgen SpaceGroupAnalyzer), while the number of sites shown in your first figure indicate the number of atoms calculated in the initial structure, in this case, a supercell.
Indeed it will be efficient to just calculate the primitive structure. But as we do high-throughput calculations, it is challenging to always guarantee that, especially for structures with experimental origin, like @Aaron_Kaplan mentioned.
Your feedback definitely will help us label these information more clearly on our website, so thank you!